pgv-simpleplot CLI Document
pgv-simpleplot is one of the CLI workflows in pyGenomeViz for simple genome visualization.
Usage
Basic Command
pgv-simpleplot --gbk_resources seq1.gbk seq2.gbk seq3.gbk seq4.gbk -o result.html
Options
General Options:
--gbk_resources IN [IN ...] Input genome genbank file resources
User can optionally specify genome range and reverse complement.
- Example1. Set 100 - 1000 range 'file:100-1000'
- Example2. Set reverse complement 'file::-1'
- Example3. Set 100 - 1000 range of reverse complement 'file:100-1000:-1'
-o OUT, --outfile OUT Output file ('*.png'|'*.jpg'|'*.svg'|'*.pdf'|'*.html')
-v, --version Print version information
-h, --help Show this help message and exit
Figure Appearence Options:
--fig_width Figure width (Default: 15)
--fig_track_height Figure track height (Default: 1.0)
--feature_track_ratio Feature track ratio (Default: 1.0)
--space_track_ratio Space (b/w feature) track ratio (Default: 2.0)
--tick_track_ratio Tick track ratio (Default: 1.0)
--track_labelsize Track label size (Default: 20)
--tick_labelsize Tick label size (Default: 15)
--align_type Figure tracks align type ('left'[*]|'center'|'right')
--tick_style Tick style ('bar'|'axis'|None[*])
--feature_plotstyle Feature plot style ('bigarrow'[*]|'arrow')
--arrow_shaft_ratio Feature arrow shaft ratio (Default: 0.5)
--feature_color Feature color (Default: 'orange')
--feature_linewidth Feature edge line width (Default: 0.0)
--dpi Figure DPI (Default: 300)
[*] marker means the default value.
Examples
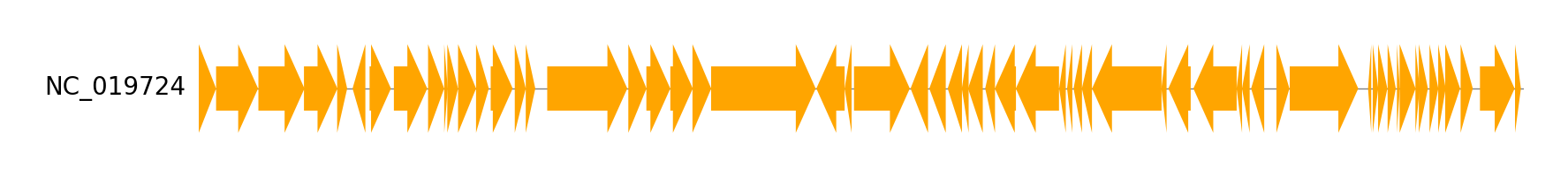
Example 1
Download example dataset:
Download four Enterobacteria phage genbank files
pgv-download-dataset -n enterobacteria_phage
Run CLI workflow:
pgv-simpleplot --gbk_resources NC_019724.gbk -o simpleplot_example1.png
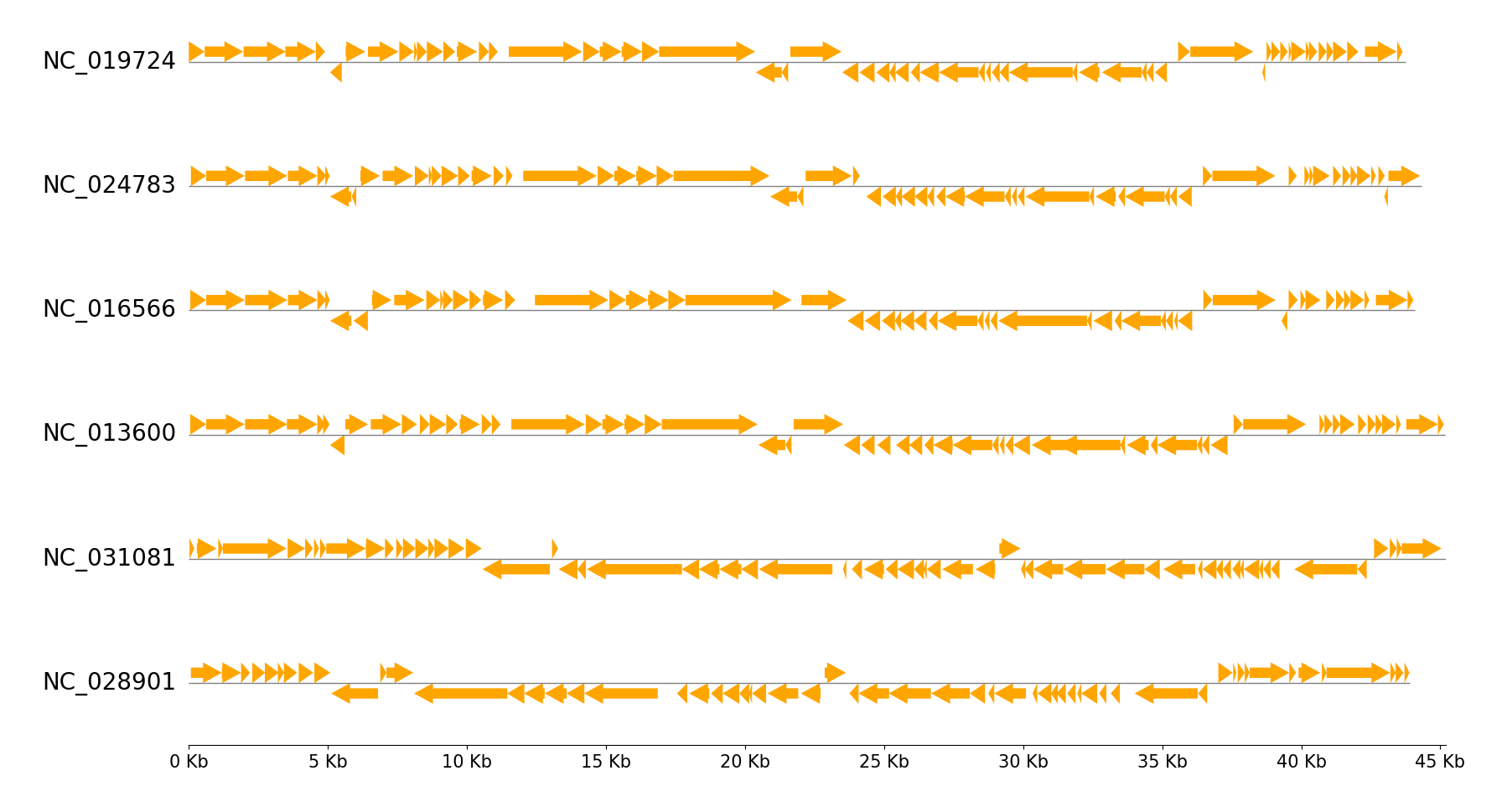
Example 2
Download six Enterobacteria phage genbank files
pgv-download-dataset -n enterobacteria_phage
Run CLI workflow:
pgv-simpleplot --gbk_resources NC_019724.gbk NC_024783.gbk NC_016566.gbk NC_013600.gbk NC_031081.gbk NC_028901.gbk \
-o simpleplot_example2.png --tick_style axis --feature_plotstyle arrow --fig_track_height 0.7

Example 3
Download example dataset:
Download four Erwinia phage genbank files
pgv-download-dataset -n erwinia_phage
Run CLI workflow:
Target range is specified (e.g. file:100-1000)
pgv-simpleplot --gbk_resources MT939486.gbk:250000-358115 MT939487.gbk:250000-355376 MT939488.gbk:250000-356948 LT960552.gbk:270000-340000 \
-o simpleplot_example3.png --tick_style bar --feature_plotstyle arrow --feature_color skyblue
