pymsaviz CLI Document
Usage
Basic Command
pymsaviz -i [MSA file] -o [MSA visualization file]
Options
$ pymsaviz --help
usage: pymsaviz [options] -i msa.fa -o msa_viz.png
MSA(Multiple Sequence Alignment) visualization CLI tool
optional arguments:
-i I, --infile I Input MSA file
-o O, --outfile O Output MSA visualization file (*.png|*.jpg|*.svg|*.pdf)
--format MSA file format (Default: 'fasta')
--color_scheme Color scheme (Default: 'Zappo')
--start Start position of MSA visualization (Default: 1)
--end End position of MSA visualization (Default: 'MSA Length')
--wrap_length Wrap length (Default: None)
--wrap_space_size Space size between wrap MSA plot area (Default: 3.0)
--label_type Label type ('id'[default]|'description')
--show_grid Show grid (Default: OFF)
--show_count Show seq char count without gap on right side (Default: OFF)
--show_consensus Show consensus sequence (Default: OFF)
--consensus_color Consensus identity bar color (Default: '#1f77b4')
--consensus_size Consensus identity bar height size (Default: 2.0)
--sort Sort MSA order by NJ tree constructed from MSA distance matrix (Default: OFF)
--dpi Figure DPI (Default: 300)
-v, --version Print version information
-h, --help Show this help message and exit
Available Color Schemes:
['Clustal', 'Zappo', 'Taylor', 'Flower', 'Blossom', 'Sunset', 'Ocean', 'Hydrophobicity', 'HelixPropensity', 'StrandPropensity', 'TurnPropensity', 'BuriedIndex', 'Nucleotide', 'Purine/Pyrimidine', 'Identity', 'None']
Example Command
Click here to download example MSA files.
Example 1
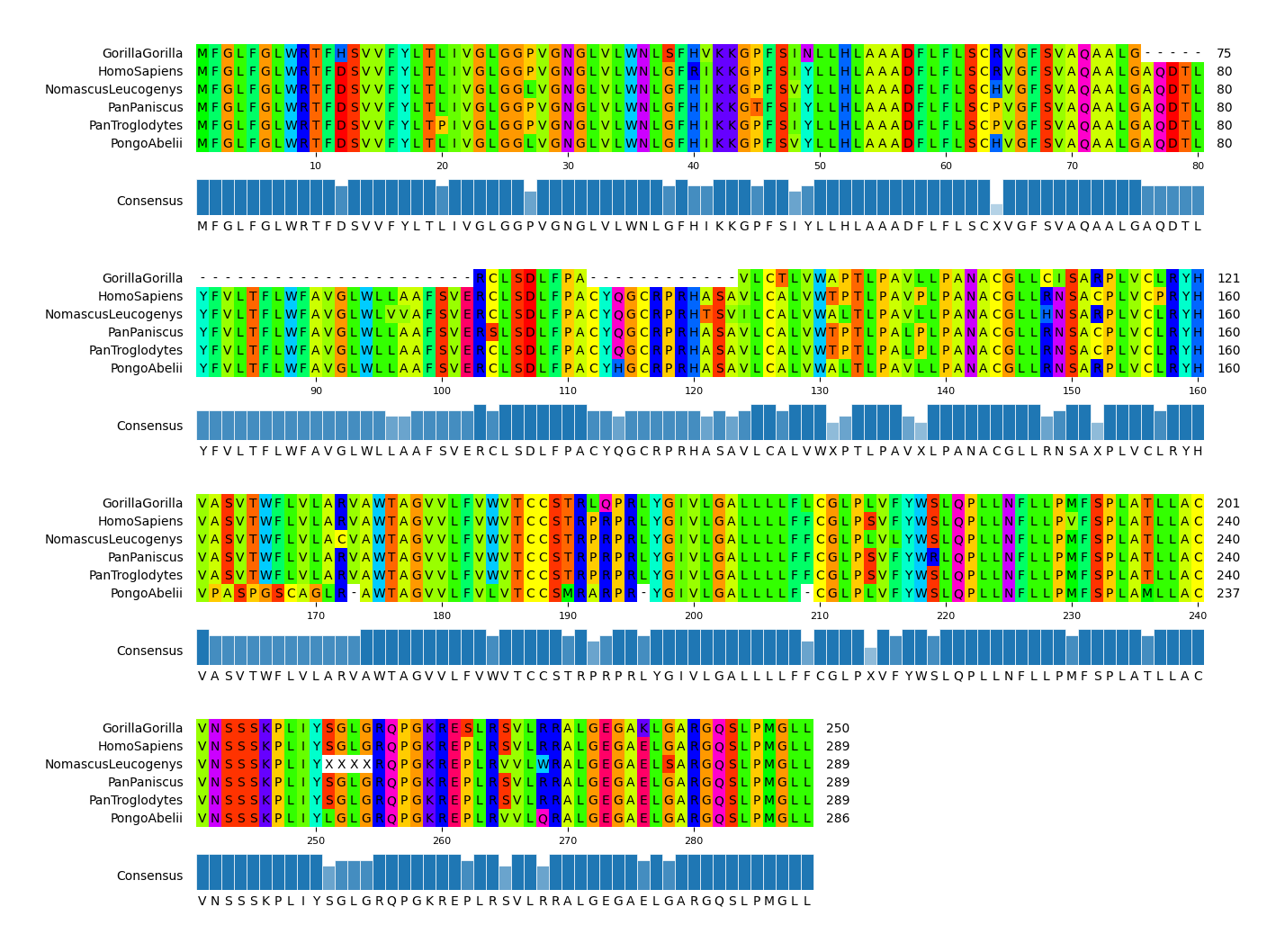
pymsaviz -i ./example/HIGD2A.fa -o cli_example01.png --color_scheme Identity
Example 2
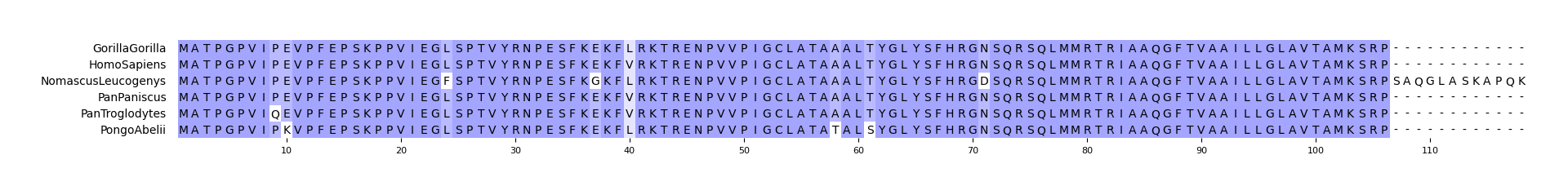
pymsaviz -i ./example/MRGPRG.fa -o cli_example02.png --wrap_length 80 \
--color_scheme Taylor --show_consensus --show_count
Example 3
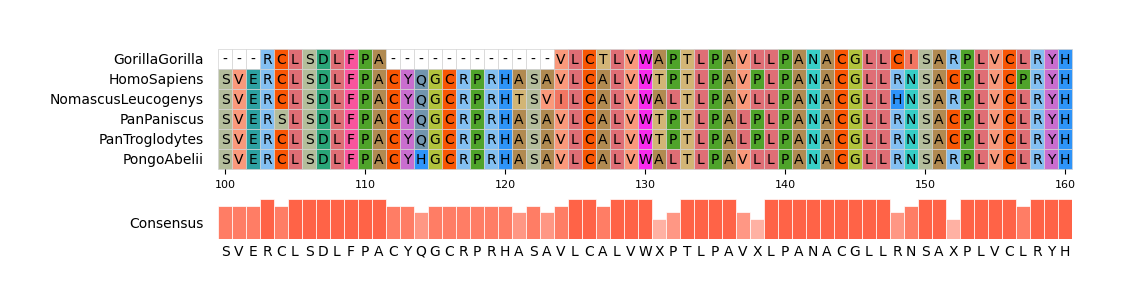
pymsaviz -i ./example/MRGPRG.fa -o cli_example03.png --start 100 --end 160 \
--color_scheme Flower --show_grid --show_consensus --consensus_color tomato